Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
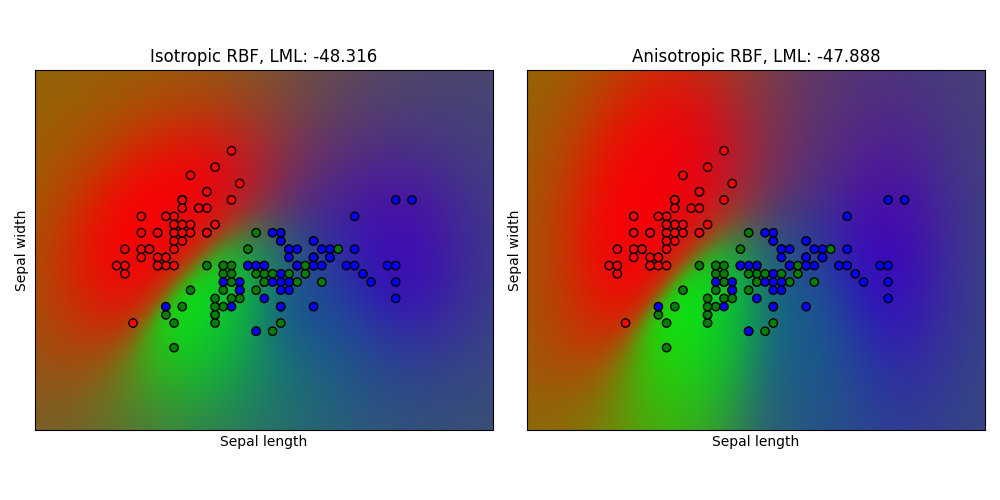
Gaussian process classification (GPC) on iris dataset#
This example illustrates the predicted probability of GPC for an isotropic and anisotropic RBF kernel on a two-dimensional version for the iris-dataset. The anisotropic RBF kernel obtains slightly higher log-marginal-likelihood by assigning different length-scales to the two feature dimensions.
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.gaussian_process.kernels import RBF
# import some data to play with
iris = datasets.load_iris()
X = iris.data[:, :2] # we only take the first two features.
y = np.array(iris.target, dtype=int)
h = 0.02 # step size in the mesh
kernel = 1.0 * RBF([1.0])
gpc_rbf_isotropic = GaussianProcessClassifier(kernel=kernel).fit(X, y)
kernel = 1.0 * RBF([1.0, 1.0])
gpc_rbf_anisotropic = GaussianProcessClassifier(kernel=kernel).fit(X, y)
# create a mesh to plot in
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
titles = ["Isotropic RBF", "Anisotropic RBF"]
plt.figure(figsize=(10, 5))
for i, clf in enumerate((gpc_rbf_isotropic, gpc_rbf_anisotropic)):
# Plot the predicted probabilities. For that, we will assign a color to
# each point in the mesh [x_min, m_max]x[y_min, y_max].
plt.subplot(1, 2, i + 1)
Z = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape((xx.shape[0], xx.shape[1], 3))
plt.imshow(Z, extent=(x_min, x_max, y_min, y_max), origin="lower")
# Plot also the training points
plt.scatter(X[:, 0], X[:, 1], c=np.array(["r", "g", "b"])[y], edgecolors=(0, 0, 0))
plt.xlabel("Sepal length")
plt.ylabel("Sepal width")
plt.xlim(xx.min(), xx.max())
plt.ylim(yy.min(), yy.max())
plt.xticks(())
plt.yticks(())
plt.title(
"%s, LML: %.3f" % (titles[i], clf.log_marginal_likelihood(clf.kernel_.theta))
)
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 3.309 seconds)
Related examples
Decision boundary of semi-supervised classifiers versus SVM on the Iris dataset
Decision boundary of semi-supervised classifiers versus SVM on the Iris dataset
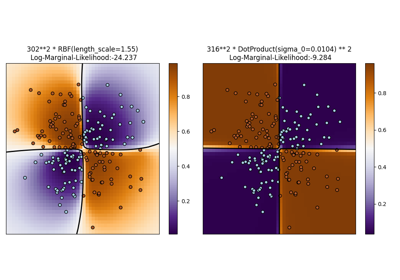
Illustration of Gaussian process classification (GPC) on the XOR dataset
Illustration of Gaussian process classification (GPC) on the XOR dataset