Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
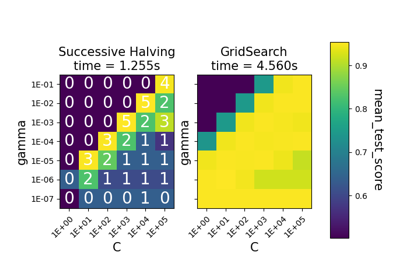
Comparing randomized search and grid search for hyperparameter estimation#
Compare randomized search and grid search for optimizing hyperparameters of a linear SVM with SGD training. All parameters that influence the learning are searched simultaneously (except for the number of estimators, which poses a time / quality tradeoff).
The randomized search and the grid search explore exactly the same space of parameters. The result in parameter settings is quite similar, while the run time for randomized search is drastically lower.
The performance is may slightly worse for the randomized search, and is likely due to a noise effect and would not carry over to a held-out test set.
Note that in practice, one would not search over this many different parameters simultaneously using grid search, but pick only the ones deemed most important.
RandomizedSearchCV took 0.85 seconds for 15 candidates parameter settings.
Model with rank: 1
Mean validation score: 0.991 (std: 0.006)
Parameters: {'alpha': 0.05063247886572012, 'average': False, 'l1_ratio': 0.13822072286080167}
Model with rank: 2
Mean validation score: 0.987 (std: 0.014)
Parameters: {'alpha': 0.010877306503748912, 'average': True, 'l1_ratio': 0.9226260871125187}
Model with rank: 3
Mean validation score: 0.976 (std: 0.023)
Parameters: {'alpha': 0.727148206404819, 'average': False, 'l1_ratio': 0.25183501383331797}
GridSearchCV took 2.82 seconds for 60 candidate parameter settings.
Model with rank: 1
Mean validation score: 0.993 (std: 0.011)
Parameters: {'alpha': 0.1, 'average': False, 'l1_ratio': 0.1111111111111111}
Model with rank: 2
Mean validation score: 0.987 (std: 0.013)
Parameters: {'alpha': 0.01, 'average': False, 'l1_ratio': 0.5555555555555556}
Model with rank: 3
Mean validation score: 0.987 (std: 0.007)
Parameters: {'alpha': 0.01, 'average': False, 'l1_ratio': 0.0}
from time import time
import numpy as np
import scipy.stats as stats
from sklearn.datasets import load_digits
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
# get some data
X, y = load_digits(return_X_y=True, n_class=3)
# build a classifier
clf = SGDClassifier(loss="hinge", penalty="elasticnet", fit_intercept=True)
# Utility function to report best scores
def report(results, n_top=3):
for i in range(1, n_top + 1):
candidates = np.flatnonzero(results["rank_test_score"] == i)
for candidate in candidates:
print("Model with rank: {0}".format(i))
print(
"Mean validation score: {0:.3f} (std: {1:.3f})".format(
results["mean_test_score"][candidate],
results["std_test_score"][candidate],
)
)
print("Parameters: {0}".format(results["params"][candidate]))
print("")
# specify parameters and distributions to sample from
param_dist = {
"average": [True, False],
"l1_ratio": stats.uniform(0, 1),
"alpha": stats.loguniform(1e-2, 1e0),
}
# run randomized search
n_iter_search = 15
random_search = RandomizedSearchCV(
clf, param_distributions=param_dist, n_iter=n_iter_search
)
start = time()
random_search.fit(X, y)
print(
"RandomizedSearchCV took %.2f seconds for %d candidates parameter settings."
% ((time() - start), n_iter_search)
)
report(random_search.cv_results_)
# use a full grid over all parameters
param_grid = {
"average": [True, False],
"l1_ratio": np.linspace(0, 1, num=10),
"alpha": np.power(10, np.arange(-2, 1, dtype=float)),
}
# run grid search
grid_search = GridSearchCV(clf, param_grid=param_grid)
start = time()
grid_search.fit(X, y)
print(
"GridSearchCV took %.2f seconds for %d candidate parameter settings."
% (time() - start, len(grid_search.cv_results_["params"]))
)
report(grid_search.cv_results_)
Total running time of the script: (0 minutes 3.678 seconds)
Related examples
Custom refit strategy of a grid search with cross-validation
Pipelining: chaining a PCA and a logistic regression
Comparison between grid search and successive halving