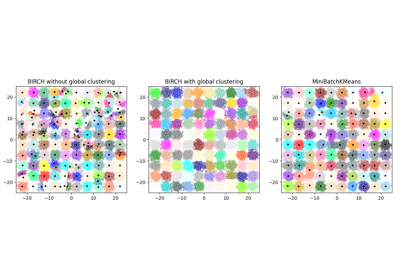
sklearn.cluster.Birch#
- class sklearn.cluster.Birch(*, threshold=0.5, branching_factor=50, n_clusters=3, compute_labels=True, copy=True)[source]#
Implements the BIRCH clustering algorithm.
It is a memory-efficient, online-learning algorithm provided as an alternative to
MiniBatchKMeans. It constructs a tree data structure with the cluster centroids being read off the leaf. These can be either the final cluster centroids or can be provided as input to another clustering algorithm such asAgglomerativeClustering.Read more in the User Guide.
New in version 0.16.
- Parameters:
- thresholdfloat, default=0.5
The radius of the subcluster obtained by merging a new sample and the closest subcluster should be lesser than the threshold. Otherwise a new subcluster is started. Setting this value to be very low promotes splitting and vice-versa.
- branching_factorint, default=50
Maximum number of CF subclusters in each node. If a new samples enters such that the number of subclusters exceed the branching_factor then that node is split into two nodes with the subclusters redistributed in each. The parent subcluster of that node is removed and two new subclusters are added as parents of the 2 split nodes.
- n_clustersint, instance of sklearn.cluster model or None, default=3
Number of clusters after the final clustering step, which treats the subclusters from the leaves as new samples.
None: the final clustering step is not performed and the subclusters are returned as they are.sklearn.clusterEstimator : If a model is provided, the model is fit treating the subclusters as new samples and the initial data is mapped to the label of the closest subcluster.int: the model fit isAgglomerativeClusteringwithn_clustersset to be equal to the int.
- compute_labelsbool, default=True
Whether or not to compute labels for each fit.
- copybool, default=True
Whether or not to make a copy of the given data. If set to False, the initial data will be overwritten.
- Attributes:
- root__CFNode
Root of the CFTree.
- dummy_leaf__CFNode
Start pointer to all the leaves.
- subcluster_centers_ndarray
Centroids of all subclusters read directly from the leaves.
- subcluster_labels_ndarray
Labels assigned to the centroids of the subclusters after they are clustered globally.
- labels_ndarray of shape (n_samples,)
Array of labels assigned to the input data. if partial_fit is used instead of fit, they are assigned to the last batch of data.
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
MiniBatchKMeansAlternative implementation that does incremental updates of the centers’ positions using mini-batches.
Notes
The tree data structure consists of nodes with each node consisting of a number of subclusters. The maximum number of subclusters in a node is determined by the branching factor. Each subcluster maintains a linear sum, squared sum and the number of samples in that subcluster. In addition, each subcluster can also have a node as its child, if the subcluster is not a member of a leaf node.
For a new point entering the root, it is merged with the subcluster closest to it and the linear sum, squared sum and the number of samples of that subcluster are updated. This is done recursively till the properties of the leaf node are updated.
References
Tian Zhang, Raghu Ramakrishnan, Maron Livny BIRCH: An efficient data clustering method for large databases. https://www.cs.sfu.ca/CourseCentral/459/han/papers/zhang96.pdf
Roberto Perdisci JBirch - Java implementation of BIRCH clustering algorithm https://code.google.com/archive/p/jbirch
Examples
>>> from sklearn.cluster import Birch >>> X = [[0, 1], [0.3, 1], [-0.3, 1], [0, -1], [0.3, -1], [-0.3, -1]] >>> brc = Birch(n_clusters=None) >>> brc.fit(X) Birch(n_clusters=None) >>> brc.predict(X) array([0, 0, 0, 1, 1, 1])
Methods
fit(X[, y])Build a CF Tree for the input data.
fit_predict(X[, y])Perform clustering on
Xand returns cluster labels.fit_transform(X[, y])Fit to data, then transform it.
get_feature_names_out([input_features])Get output feature names for transformation.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
partial_fit([X, y])Online learning.
predict(X)Predict data using the
centroids_of subclusters.set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform X into subcluster centroids dimension.
- fit(X, y=None)[source]#
Build a CF Tree for the input data.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Input data.
- yIgnored
Not used, present here for API consistency by convention.
- Returns:
- self
Fitted estimator.
- fit_predict(X, y=None, **kwargs)[source]#
Perform clustering on
Xand returns cluster labels.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input data.
- yIgnored
Not used, present for API consistency by convention.
- **kwargsdict
Arguments to be passed to
fit.New in version 1.4.
- Returns:
- labelsndarray of shape (n_samples,), dtype=np.int64
Cluster labels.
- fit_transform(X, y=None, **fit_params)[source]#
Fit to data, then transform it.
Fits transformer to
Xandywith optional parametersfit_paramsand returns a transformed version ofX.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs), default=None
Target values (None for unsupervised transformations).
- **fit_paramsdict
Additional fit parameters.
- Returns:
- X_newndarray array of shape (n_samples, n_features_new)
Transformed array.
- get_feature_names_out(input_features=None)[source]#
Get output feature names for transformation.
The feature names out will prefixed by the lowercased class name. For example, if the transformer outputs 3 features, then the feature names out are:
["class_name0", "class_name1", "class_name2"].- Parameters:
- input_featuresarray-like of str or None, default=None
Only used to validate feature names with the names seen in
fit.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- partial_fit(X=None, y=None)[source]#
Online learning. Prevents rebuilding of CFTree from scratch.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features), default=None
Input data. If X is not provided, only the global clustering step is done.
- yIgnored
Not used, present here for API consistency by convention.
- Returns:
- self
Fitted estimator.
- predict(X)[source]#
Predict data using the
centroids_of subclusters.Avoid computation of the row norms of X.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Input data.
- Returns:
- labelsndarray of shape(n_samples,)
Labelled data.
- set_output(*, transform=None)[source]#
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame output"polars": Polars outputNone: Transform configuration is unchanged
New in version 1.4:
"polars"option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- transform(X)[source]#
Transform X into subcluster centroids dimension.
Each dimension represents the distance from the sample point to each cluster centroid.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Input data.
- Returns:
- X_trans{array-like, sparse matrix} of shape (n_samples, n_clusters)
Transformed data.
Examples using sklearn.cluster.Birch#
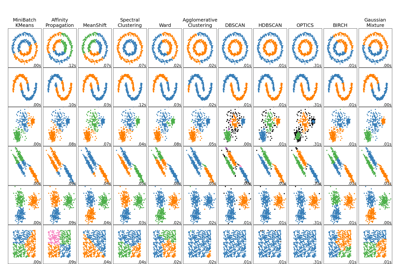
Comparing different clustering algorithms on toy datasets