sklearn.svm.LinearSVC#
- class sklearn.svm.LinearSVC(penalty='l2', loss='squared_hinge', *, dual='warn', tol=0.0001, C=1.0, multi_class='ovr', fit_intercept=True, intercept_scaling=1, class_weight=None, verbose=0, random_state=None, max_iter=1000)[source]#
Linear Support Vector Classification.
Similar to SVC with parameter kernel=’linear’, but implemented in terms of liblinear rather than libsvm, so it has more flexibility in the choice of penalties and loss functions and should scale better to large numbers of samples.
The main differences between
LinearSVCandSVClie in the loss function used by default, and in the handling of intercept regularization between those two implementations.This class supports both dense and sparse input and the multiclass support is handled according to a one-vs-the-rest scheme.
Read more in the User Guide.
- Parameters:
- penalty{‘l1’, ‘l2’}, default=’l2’
Specifies the norm used in the penalization. The ‘l2’ penalty is the standard used in SVC. The ‘l1’ leads to
coef_vectors that are sparse.- loss{‘hinge’, ‘squared_hinge’}, default=’squared_hinge’
Specifies the loss function. ‘hinge’ is the standard SVM loss (used e.g. by the SVC class) while ‘squared_hinge’ is the square of the hinge loss. The combination of
penalty='l1'andloss='hinge'is not supported.- dual“auto” or bool, default=True
Select the algorithm to either solve the dual or primal optimization problem. Prefer dual=False when n_samples > n_features.
dual="auto"will choose the value of the parameter automatically, based on the values ofn_samples,n_features,loss,multi_classandpenalty. Ifn_samples<n_featuresand optimizer supports chosenloss,multi_classandpenalty, then dual will be set to True, otherwise it will be set to False.Changed in version 1.3: The
"auto"option is added in version 1.3 and will be the default in version 1.5.- tolfloat, default=1e-4
Tolerance for stopping criteria.
- Cfloat, default=1.0
Regularization parameter. The strength of the regularization is inversely proportional to C. Must be strictly positive.
- multi_class{‘ovr’, ‘crammer_singer’}, default=’ovr’
Determines the multi-class strategy if
ycontains more than two classes."ovr"trains n_classes one-vs-rest classifiers, while"crammer_singer"optimizes a joint objective over all classes. Whilecrammer_singeris interesting from a theoretical perspective as it is consistent, it is seldom used in practice as it rarely leads to better accuracy and is more expensive to compute. If"crammer_singer"is chosen, the options loss, penalty and dual will be ignored.- fit_interceptbool, default=True
Whether or not to fit an intercept. If set to True, the feature vector is extended to include an intercept term:
[x_1, ..., x_n, 1], where 1 corresponds to the intercept. If set to False, no intercept will be used in calculations (i.e. data is expected to be already centered).- intercept_scalingfloat, default=1.0
When
fit_interceptis True, the instance vector x becomes[x_1, ..., x_n, intercept_scaling], i.e. a “synthetic” feature with a constant value equal tointercept_scalingis appended to the instance vector. The intercept becomes intercept_scaling * synthetic feature weight. Note that liblinear internally penalizes the intercept, treating it like any other term in the feature vector. To reduce the impact of the regularization on the intercept, theintercept_scalingparameter can be set to a value greater than 1; the higher the value ofintercept_scaling, the lower the impact of regularization on it. Then, the weights become[w_x_1, ..., w_x_n, w_intercept*intercept_scaling], wherew_x_1, ..., w_x_nrepresent the feature weights and the intercept weight is scaled byintercept_scaling. This scaling allows the intercept term to have a different regularization behavior compared to the other features.- class_weightdict or ‘balanced’, default=None
Set the parameter C of class i to
class_weight[i]*Cfor SVC. If not given, all classes are supposed to have weight one. The “balanced” mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data asn_samples / (n_classes * np.bincount(y)).- verboseint, default=0
Enable verbose output. Note that this setting takes advantage of a per-process runtime setting in liblinear that, if enabled, may not work properly in a multithreaded context.
- random_stateint, RandomState instance or None, default=None
Controls the pseudo random number generation for shuffling the data for the dual coordinate descent (if
dual=True). Whendual=Falsethe underlying implementation ofLinearSVCis not random andrandom_statehas no effect on the results. Pass an int for reproducible output across multiple function calls. See Glossary.- max_iterint, default=1000
The maximum number of iterations to be run.
- Attributes:
- coef_ndarray of shape (1, n_features) if n_classes == 2 else (n_classes, n_features)
Weights assigned to the features (coefficients in the primal problem).
coef_is a readonly property derived fromraw_coef_that follows the internal memory layout of liblinear.- intercept_ndarray of shape (1,) if n_classes == 2 else (n_classes,)
Constants in decision function.
- classes_ndarray of shape (n_classes,)
The unique classes labels.
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
- n_iter_int
Maximum number of iterations run across all classes.
See also
SVCImplementation of Support Vector Machine classifier using libsvm: the kernel can be non-linear but its SMO algorithm does not scale to large number of samples as LinearSVC does. Furthermore SVC multi-class mode is implemented using one vs one scheme while LinearSVC uses one vs the rest. It is possible to implement one vs the rest with SVC by using the
OneVsRestClassifierwrapper. Finally SVC can fit dense data without memory copy if the input is C-contiguous. Sparse data will still incur memory copy though.sklearn.linear_model.SGDClassifierSGDClassifier can optimize the same cost function as LinearSVC by adjusting the penalty and loss parameters. In addition it requires less memory, allows incremental (online) learning, and implements various loss functions and regularization regimes.
Notes
The underlying C implementation uses a random number generator to select features when fitting the model. It is thus not uncommon to have slightly different results for the same input data. If that happens, try with a smaller
tolparameter.The underlying implementation, liblinear, uses a sparse internal representation for the data that will incur a memory copy.
Predict output may not match that of standalone liblinear in certain cases. See differences from liblinear in the narrative documentation.
References
LIBLINEAR: A Library for Large Linear Classification
Examples
>>> from sklearn.svm import LinearSVC >>> from sklearn.pipeline import make_pipeline >>> from sklearn.preprocessing import StandardScaler >>> from sklearn.datasets import make_classification >>> X, y = make_classification(n_features=4, random_state=0) >>> clf = make_pipeline(StandardScaler(), ... LinearSVC(dual="auto", random_state=0, tol=1e-5)) >>> clf.fit(X, y) Pipeline(steps=[('standardscaler', StandardScaler()), ('linearsvc', LinearSVC(dual='auto', random_state=0, tol=1e-05))])
>>> print(clf.named_steps['linearsvc'].coef_) [[0.141... 0.526... 0.679... 0.493...]]
>>> print(clf.named_steps['linearsvc'].intercept_) [0.1693...] >>> print(clf.predict([[0, 0, 0, 0]])) [1]
Methods
Predict confidence scores for samples.
densify()Convert coefficient matrix to dense array format.
fit(X, y[, sample_weight])Fit the model according to the given training data.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
predict(X)Predict class labels for samples in X.
score(X, y[, sample_weight])Return the mean accuracy on the given test data and labels.
set_fit_request(*[, sample_weight])Request metadata passed to the
fitmethod.set_params(**params)Set the parameters of this estimator.
set_score_request(*[, sample_weight])Request metadata passed to the
scoremethod.sparsify()Convert coefficient matrix to sparse format.
- decision_function(X)[source]#
Predict confidence scores for samples.
The confidence score for a sample is proportional to the signed distance of that sample to the hyperplane.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The data matrix for which we want to get the confidence scores.
- Returns:
- scoresndarray of shape (n_samples,) or (n_samples, n_classes)
Confidence scores per
(n_samples, n_classes)combination. In the binary case, confidence score forself.classes_[1]where >0 means this class would be predicted.
- densify()[source]#
Convert coefficient matrix to dense array format.
Converts the
coef_member (back) to a numpy.ndarray. This is the default format ofcoef_and is required for fitting, so calling this method is only required on models that have previously been sparsified; otherwise, it is a no-op.- Returns:
- self
Fitted estimator.
- fit(X, y, sample_weight=None)[source]#
Fit the model according to the given training data.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Training vector, where
n_samplesis the number of samples andn_featuresis the number of features.- yarray-like of shape (n_samples,)
Target vector relative to X.
- sample_weightarray-like of shape (n_samples,), default=None
Array of weights that are assigned to individual samples. If not provided, then each sample is given unit weight.
New in version 0.18.
- Returns:
- selfobject
An instance of the estimator.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- predict(X)[source]#
Predict class labels for samples in X.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The data matrix for which we want to get the predictions.
- Returns:
- y_predndarray of shape (n_samples,)
Vector containing the class labels for each sample.
- score(X, y, sample_weight=None)[source]#
Return the mean accuracy on the given test data and labels.
In multi-label classification, this is the subset accuracy which is a harsh metric since you require for each sample that each label set be correctly predicted.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Test samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True labels for
X.- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns:
- scorefloat
Mean accuracy of
self.predict(X)w.r.t.y.
- set_fit_request(*, sample_weight: bool | None | str = '$UNCHANGED$') LinearSVC[source]#
Request metadata passed to the
fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
Pipeline. Otherwise it has no effect.- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- set_score_request(*, sample_weight: bool | None | str = '$UNCHANGED$') LinearSVC[source]#
Request metadata passed to the
scoremethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toscoreif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toscore.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
Pipeline. Otherwise it has no effect.- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter inscore.
- Returns:
- selfobject
The updated object.
- sparsify()[source]#
Convert coefficient matrix to sparse format.
Converts the
coef_member to a scipy.sparse matrix, which for L1-regularized models can be much more memory- and storage-efficient than the usual numpy.ndarray representation.The
intercept_member is not converted.- Returns:
- self
Fitted estimator.
Notes
For non-sparse models, i.e. when there are not many zeros in
coef_, this may actually increase memory usage, so use this method with care. A rule of thumb is that the number of zero elements, which can be computed with(coef_ == 0).sum(), must be more than 50% for this to provide significant benefits.After calling this method, further fitting with the partial_fit method (if any) will not work until you call densify.
Examples using sklearn.svm.LinearSVC#
Scalable learning with polynomial kernel approximation
Explicit feature map approximation for RBF kernels
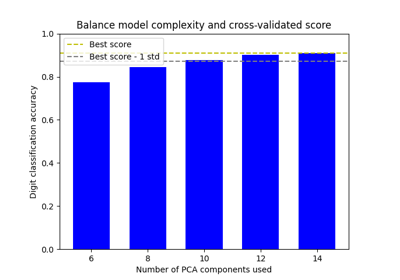
Balance model complexity and cross-validated score
Column Transformer with Heterogeneous Data Sources
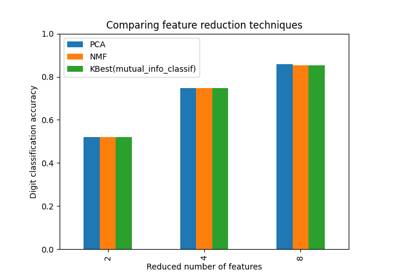
Selecting dimensionality reduction with Pipeline and GridSearchCV
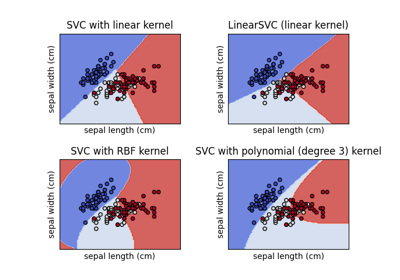
Plot different SVM classifiers in the iris dataset

Classification of text documents using sparse features