sklearn.ensemble.RandomTreesEmbedding#
- class sklearn.ensemble.RandomTreesEmbedding(n_estimators=100, *, max_depth=5, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_leaf_nodes=None, min_impurity_decrease=0.0, sparse_output=True, n_jobs=None, random_state=None, verbose=0, warm_start=False)[source]#
An ensemble of totally random trees.
An unsupervised transformation of a dataset to a high-dimensional sparse representation. A datapoint is coded according to which leaf of each tree it is sorted into. Using a one-hot encoding of the leaves, this leads to a binary coding with as many ones as there are trees in the forest.
The dimensionality of the resulting representation is
n_out <= n_estimators * max_leaf_nodes. Ifmax_leaf_nodes == None, the number of leaf nodes is at mostn_estimators * 2 ** max_depth.Read more in the User Guide.
- Parameters:
- n_estimatorsint, default=100
Number of trees in the forest.
Changed in version 0.22: The default value of
n_estimatorschanged from 10 to 100 in 0.22.- max_depthint, default=5
The maximum depth of each tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples.
- min_samples_splitint or float, default=2
The minimum number of samples required to split an internal node:
If int, then consider
min_samples_splitas the minimum number.If float, then
min_samples_splitis a fraction andceil(min_samples_split * n_samples)is the minimum number of samples for each split.
Changed in version 0.18: Added float values for fractions.
- min_samples_leafint or float, default=1
The minimum number of samples required to be at a leaf node. A split point at any depth will only be considered if it leaves at least
min_samples_leaftraining samples in each of the left and right branches. This may have the effect of smoothing the model, especially in regression.If int, then consider
min_samples_leafas the minimum number.If float, then
min_samples_leafis a fraction andceil(min_samples_leaf * n_samples)is the minimum number of samples for each node.
Changed in version 0.18: Added float values for fractions.
- min_weight_fraction_leaffloat, default=0.0
The minimum weighted fraction of the sum total of weights (of all the input samples) required to be at a leaf node. Samples have equal weight when sample_weight is not provided.
- max_leaf_nodesint, default=None
Grow trees with
max_leaf_nodesin best-first fashion. Best nodes are defined as relative reduction in impurity. If None then unlimited number of leaf nodes.- min_impurity_decreasefloat, default=0.0
A node will be split if this split induces a decrease of the impurity greater than or equal to this value.
The weighted impurity decrease equation is the following:
N_t / N * (impurity - N_t_R / N_t * right_impurity - N_t_L / N_t * left_impurity)
where
Nis the total number of samples,N_tis the number of samples at the current node,N_t_Lis the number of samples in the left child, andN_t_Ris the number of samples in the right child.N,N_t,N_t_RandN_t_Lall refer to the weighted sum, ifsample_weightis passed.New in version 0.19.
- sparse_outputbool, default=True
Whether or not to return a sparse CSR matrix, as default behavior, or to return a dense array compatible with dense pipeline operators.
- n_jobsint, default=None
The number of jobs to run in parallel.
fit,transform,decision_pathandapplyare all parallelized over the trees.Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- random_stateint, RandomState instance or None, default=None
Controls the generation of the random
yused to fit the trees and the draw of the splits for each feature at the trees’ nodes. See Glossary for details.- verboseint, default=0
Controls the verbosity when fitting and predicting.
- warm_startbool, default=False
When set to
True, reuse the solution of the previous call to fit and add more estimators to the ensemble, otherwise, just fit a whole new forest. See Glossary and Fitting additional weak-learners for details.
- Attributes:
- estimator_
ExtraTreeRegressorinstance The child estimator template used to create the collection of fitted sub-estimators.
New in version 1.2:
base_estimator_was renamed toestimator_.- estimators_list of
ExtraTreeRegressorinstances The collection of fitted sub-estimators.
feature_importances_ndarray of shape (n_features,)The impurity-based feature importances.
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
- n_outputs_int
The number of outputs when
fitis performed.- one_hot_encoder_OneHotEncoder instance
One-hot encoder used to create the sparse embedding.
estimators_samples_list of arraysThe subset of drawn samples for each base estimator.
- estimator_
See also
ExtraTreesClassifierAn extra-trees classifier.
ExtraTreesRegressorAn extra-trees regressor.
RandomForestClassifierA random forest classifier.
RandomForestRegressorA random forest regressor.
sklearn.tree.ExtraTreeClassifierAn extremely randomized tree classifier.
sklearn.tree.ExtraTreeRegressorAn extremely randomized tree regressor.
References
[1]P. Geurts, D. Ernst., and L. Wehenkel, “Extremely randomized trees”, Machine Learning, 63(1), 3-42, 2006.
[2]Moosmann, F. and Triggs, B. and Jurie, F. “Fast discriminative visual codebooks using randomized clustering forests” NIPS 2007
Examples
>>> from sklearn.ensemble import RandomTreesEmbedding >>> X = [[0,0], [1,0], [0,1], [-1,0], [0,-1]] >>> random_trees = RandomTreesEmbedding( ... n_estimators=5, random_state=0, max_depth=1).fit(X) >>> X_sparse_embedding = random_trees.transform(X) >>> X_sparse_embedding.toarray() array([[0., 1., 1., 0., 1., 0., 0., 1., 1., 0.], [0., 1., 1., 0., 1., 0., 0., 1., 1., 0.], [0., 1., 0., 1., 0., 1., 0., 1., 0., 1.], [1., 0., 1., 0., 1., 0., 1., 0., 1., 0.], [0., 1., 1., 0., 1., 0., 0., 1., 1., 0.]])
Methods
apply(X)Apply trees in the forest to X, return leaf indices.
Return the decision path in the forest.
fit(X[, y, sample_weight])Fit estimator.
fit_transform(X[, y, sample_weight])Fit estimator and transform dataset.
get_feature_names_out([input_features])Get output feature names for transformation.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
set_fit_request(*[, sample_weight])Request metadata passed to the
fitmethod.set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform dataset.
- apply(X)[source]#
Apply trees in the forest to X, return leaf indices.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The input samples. Internally, its dtype will be converted to
dtype=np.float32. If a sparse matrix is provided, it will be converted into a sparsecsr_matrix.
- Returns:
- X_leavesndarray of shape (n_samples, n_estimators)
For each datapoint x in X and for each tree in the forest, return the index of the leaf x ends up in.
- decision_path(X)[source]#
Return the decision path in the forest.
New in version 0.18.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The input samples. Internally, its dtype will be converted to
dtype=np.float32. If a sparse matrix is provided, it will be converted into a sparsecsr_matrix.
- Returns:
- indicatorsparse matrix of shape (n_samples, n_nodes)
Return a node indicator matrix where non zero elements indicates that the samples goes through the nodes. The matrix is of CSR format.
- n_nodes_ptrndarray of shape (n_estimators + 1,)
The columns from indicator[n_nodes_ptr[i]:n_nodes_ptr[i+1]] gives the indicator value for the i-th estimator.
- property estimators_samples_#
The subset of drawn samples for each base estimator.
Returns a dynamically generated list of indices identifying the samples used for fitting each member of the ensemble, i.e., the in-bag samples.
Note: the list is re-created at each call to the property in order to reduce the object memory footprint by not storing the sampling data. Thus fetching the property may be slower than expected.
- property feature_importances_#
The impurity-based feature importances.
The higher, the more important the feature. The importance of a feature is computed as the (normalized) total reduction of the criterion brought by that feature. It is also known as the Gini importance.
Warning: impurity-based feature importances can be misleading for high cardinality features (many unique values). See
sklearn.inspection.permutation_importanceas an alternative.- Returns:
- feature_importances_ndarray of shape (n_features,)
The values of this array sum to 1, unless all trees are single node trees consisting of only the root node, in which case it will be an array of zeros.
- fit(X, y=None, sample_weight=None)[source]#
Fit estimator.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The input samples. Use
dtype=np.float32for maximum efficiency. Sparse matrices are also supported, use sparsecsc_matrixfor maximum efficiency.- yIgnored
Not used, present for API consistency by convention.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights. If None, then samples are equally weighted. Splits that would create child nodes with net zero or negative weight are ignored while searching for a split in each node. In the case of classification, splits are also ignored if they would result in any single class carrying a negative weight in either child node.
- Returns:
- selfobject
Returns the instance itself.
- fit_transform(X, y=None, sample_weight=None)[source]#
Fit estimator and transform dataset.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Input data used to build forests. Use
dtype=np.float32for maximum efficiency.- yIgnored
Not used, present for API consistency by convention.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights. If None, then samples are equally weighted. Splits that would create child nodes with net zero or negative weight are ignored while searching for a split in each node. In the case of classification, splits are also ignored if they would result in any single class carrying a negative weight in either child node.
- Returns:
- X_transformedsparse matrix of shape (n_samples, n_out)
Transformed dataset.
- get_feature_names_out(input_features=None)[source]#
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Only used to validate feature names with the names seen in
fit.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names, in the format of
randomtreesembedding_{tree}_{leaf}, wheretreeis the tree used to generate the leaf andleafis the index of a leaf node in that tree. Note that the node indexing scheme is used to index both nodes with children (split nodes) and leaf nodes. Only the latter can be present as output features. As a consequence, there are missing indices in the output feature names.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- set_fit_request(*, sample_weight: bool | None | str = '$UNCHANGED$') RandomTreesEmbedding[source]#
Request metadata passed to the
fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
Pipeline. Otherwise it has no effect.- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.
- Returns:
- selfobject
The updated object.
- set_output(*, transform=None)[source]#
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame output"polars": Polars outputNone: Transform configuration is unchanged
New in version 1.4:
"polars"option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- transform(X)[source]#
Transform dataset.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Input data to be transformed. Use
dtype=np.float32for maximum efficiency. Sparse matrices are also supported, use sparsecsr_matrixfor maximum efficiency.
- Returns:
- X_transformedsparse matrix of shape (n_samples, n_out)
Transformed dataset.
Examples using sklearn.ensemble.RandomTreesEmbedding#
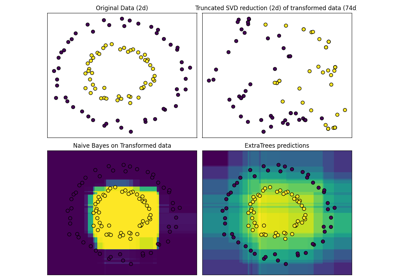
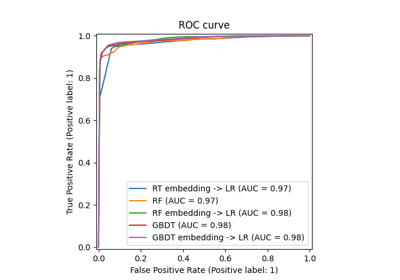
Hashing feature transformation using Totally Random Trees
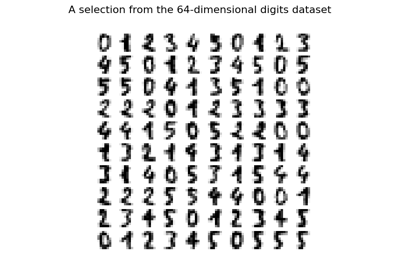
Manifold learning on handwritten digits: Locally Linear Embedding, Isomap…