sklearn.compose.ColumnTransformer#
- class sklearn.compose.ColumnTransformer(transformers, *, remainder='drop', sparse_threshold=0.3, n_jobs=None, transformer_weights=None, verbose=False, verbose_feature_names_out=True)[source]#
Applies transformers to columns of an array or pandas DataFrame.
This estimator allows different columns or column subsets of the input to be transformed separately and the features generated by each transformer will be concatenated to form a single feature space. This is useful for heterogeneous or columnar data, to combine several feature extraction mechanisms or transformations into a single transformer.
Read more in the User Guide.
New in version 0.20.
- Parameters:
- transformerslist of tuples
List of (name, transformer, columns) tuples specifying the transformer objects to be applied to subsets of the data.
- namestr
Like in Pipeline and FeatureUnion, this allows the transformer and its parameters to be set using
set_paramsand searched in grid search.- transformer{‘drop’, ‘passthrough’} or estimator
Estimator must support fit and transform. Special-cased strings ‘drop’ and ‘passthrough’ are accepted as well, to indicate to drop the columns or to pass them through untransformed, respectively.
- columnsstr, array-like of str, int, array-like of int, array-like of bool, slice or callable
Indexes the data on its second axis. Integers are interpreted as positional columns, while strings can reference DataFrame columns by name. A scalar string or int should be used where
transformerexpects X to be a 1d array-like (vector), otherwise a 2d array will be passed to the transformer. A callable is passed the input dataXand can return any of the above. To select multiple columns by name or dtype, you can usemake_column_selector.
- remainder{‘drop’, ‘passthrough’} or estimator, default=’drop’
By default, only the specified columns in
transformersare transformed and combined in the output, and the non-specified columns are dropped. (default of'drop'). By specifyingremainder='passthrough', all remaining columns that were not specified intransformers, but present in the data passed tofitwill be automatically passed through. This subset of columns is concatenated with the output of the transformers. For dataframes, extra columns not seen duringfitwill be excluded from the output oftransform. By settingremainderto be an estimator, the remaining non-specified columns will use theremainderestimator. The estimator must support fit and transform. Note that using this feature requires that the DataFrame columns input at fit and transform have identical order.- sparse_thresholdfloat, default=0.3
If the output of the different transformers contains sparse matrices, these will be stacked as a sparse matrix if the overall density is lower than this value. Use
sparse_threshold=0to always return dense. When the transformed output consists of all dense data, the stacked result will be dense, and this keyword will be ignored.- n_jobsint, default=None
Number of jobs to run in parallel.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- transformer_weightsdict, default=None
Multiplicative weights for features per transformer. The output of the transformer is multiplied by these weights. Keys are transformer names, values the weights.
- verbosebool, default=False
If True, the time elapsed while fitting each transformer will be printed as it is completed.
- verbose_feature_names_outbool, default=True
If True,
ColumnTransformer.get_feature_names_outwill prefix all feature names with the name of the transformer that generated that feature. If False,ColumnTransformer.get_feature_names_outwill not prefix any feature names and will error if feature names are not unique.New in version 1.0.
- Attributes:
- transformers_list
The collection of fitted transformers as tuples of (name, fitted_transformer, column).
fitted_transformercan be an estimator, or'drop';'passthrough'is replaced with an equivalentFunctionTransformer. In case there were no columns selected, this will be the unfitted transformer. If there are remaining columns, the final element is a tuple of the form: (‘remainder’, transformer, remaining_columns) corresponding to theremainderparameter. If there are remaining columns, thenlen(transformers_)==len(transformers)+1, otherwiselen(transformers_)==len(transformers).named_transformers_BunchAccess the fitted transformer by name.
- sparse_output_bool
Boolean flag indicating whether the output of
transformis a sparse matrix or a dense numpy array, which depends on the output of the individual transformers and thesparse_thresholdkeyword.- output_indices_dict
A dictionary from each transformer name to a slice, where the slice corresponds to indices in the transformed output. This is useful to inspect which transformer is responsible for which transformed feature(s).
New in version 1.0.
- n_features_in_int
Number of features seen during fit. Only defined if the underlying transformers expose such an attribute when fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
make_column_transformerConvenience function for combining the outputs of multiple transformer objects applied to column subsets of the original feature space.
make_column_selectorConvenience function for selecting columns based on datatype or the columns name with a regex pattern.
Notes
The order of the columns in the transformed feature matrix follows the order of how the columns are specified in the
transformerslist. Columns of the original feature matrix that are not specified are dropped from the resulting transformed feature matrix, unless specified in thepassthroughkeyword. Those columns specified withpassthroughare added at the right to the output of the transformers.Examples
>>> import numpy as np >>> from sklearn.compose import ColumnTransformer >>> from sklearn.preprocessing import Normalizer >>> ct = ColumnTransformer( ... [("norm1", Normalizer(norm='l1'), [0, 1]), ... ("norm2", Normalizer(norm='l1'), slice(2, 4))]) >>> X = np.array([[0., 1., 2., 2.], ... [1., 1., 0., 1.]]) >>> # Normalizer scales each row of X to unit norm. A separate scaling >>> # is applied for the two first and two last elements of each >>> # row independently. >>> ct.fit_transform(X) array([[0. , 1. , 0.5, 0.5], [0.5, 0.5, 0. , 1. ]])
ColumnTransformercan be configured with a transformer that requires a 1d array by setting the column to a string:>>> from sklearn.feature_extraction import FeatureHasher >>> from sklearn.preprocessing import MinMaxScaler >>> import pandas as pd >>> X = pd.DataFrame({ ... "documents": ["First item", "second one here", "Is this the last?"], ... "width": [3, 4, 5], ... }) >>> # "documents" is a string which configures ColumnTransformer to >>> # pass the documents column as a 1d array to the FeatureHasher >>> ct = ColumnTransformer( ... [("text_preprocess", FeatureHasher(input_type="string"), "documents"), ... ("num_preprocess", MinMaxScaler(), ["width"])]) >>> X_trans = ct.fit_transform(X)
For a more detailed example of usage, see Column Transformer with Mixed Types.
Methods
fit(X[, y])Fit all transformers using X.
fit_transform(X[, y])Fit all transformers, transform the data and concatenate results.
get_feature_names_out([input_features])Get output feature names for transformation.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
set_output(*[, transform])Set the output container when
"transform"and"fit_transform"are called.set_params(**kwargs)Set the parameters of this estimator.
transform(X, **params)Transform X separately by each transformer, concatenate results.
- fit(X, y=None, **params)[source]#
Fit all transformers using X.
- Parameters:
- X{array-like, dataframe} of shape (n_samples, n_features)
Input data, of which specified subsets are used to fit the transformers.
- yarray-like of shape (n_samples,…), default=None
Targets for supervised learning.
- **paramsdict, default=None
Parameters to be passed to the underlying transformers’
fitandtransformmethods.You can only pass this if metadata routing is enabled, which you can enable using
sklearn.set_config(enable_metadata_routing=True).New in version 1.4.
- Returns:
- selfColumnTransformer
This estimator.
- fit_transform(X, y=None, **params)[source]#
Fit all transformers, transform the data and concatenate results.
- Parameters:
- X{array-like, dataframe} of shape (n_samples, n_features)
Input data, of which specified subsets are used to fit the transformers.
- yarray-like of shape (n_samples,), default=None
Targets for supervised learning.
- **paramsdict, default=None
Parameters to be passed to the underlying transformers’
fitandtransformmethods.You can only pass this if metadata routing is enabled, which you can enable using
sklearn.set_config(enable_metadata_routing=True).New in version 1.4.
- Returns:
- X_t{array-like, sparse matrix} of shape (n_samples, sum_n_components)
Horizontally stacked results of transformers. sum_n_components is the sum of n_components (output dimension) over transformers. If any result is a sparse matrix, everything will be converted to sparse matrices.
- get_feature_names_out(input_features=None)[source]#
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
New in version 1.4.
- Returns:
- routingMetadataRouter
A
MetadataRouterencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
Returns the parameters given in the constructor as well as the estimators contained within the
transformersof theColumnTransformer.- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- property named_transformers_#
Access the fitted transformer by name.
Read-only attribute to access any transformer by given name. Keys are transformer names and values are the fitted transformer objects.
- set_output(*, transform=None)[source]#
Set the output container when
"transform"and"fit_transform"are called.Calling
set_outputwill set the output of all estimators intransformersandtransformers_.- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame output"polars": Polars outputNone: Transform configuration is unchanged
New in version 1.4:
"polars"option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**kwargs)[source]#
Set the parameters of this estimator.
Valid parameter keys can be listed with
get_params(). Note that you can directly set the parameters of the estimators contained intransformersofColumnTransformer.- Parameters:
- **kwargsdict
Estimator parameters.
- Returns:
- selfColumnTransformer
This estimator.
- transform(X, **params)[source]#
Transform X separately by each transformer, concatenate results.
- Parameters:
- X{array-like, dataframe} of shape (n_samples, n_features)
The data to be transformed by subset.
- **paramsdict, default=None
Parameters to be passed to the underlying transformers’
transformmethod.You can only pass this if metadata routing is enabled, which you can enable using
sklearn.set_config(enable_metadata_routing=True).New in version 1.4.
- Returns:
- X_t{array-like, sparse matrix} of shape (n_samples, sum_n_components)
Horizontally stacked results of transformers. sum_n_components is the sum of n_components (output dimension) over transformers. If any result is a sparse matrix, everything will be converted to sparse matrices.
Examples using sklearn.compose.ColumnTransformer#
Partial Dependence and Individual Conditional Expectation Plots
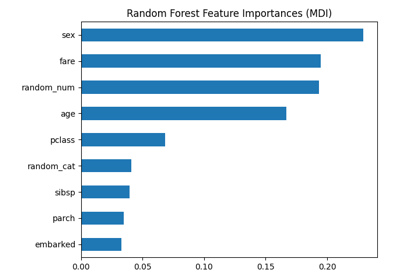
Permutation Importance vs Random Forest Feature Importance (MDI)
Column Transformer with Heterogeneous Data Sources