sklearn.model_selection.cross_val_score#
- sklearn.model_selection.cross_val_score(estimator, X, y=None, *, groups=None, scoring=None, cv=None, n_jobs=None, verbose=0, fit_params=None, params=None, pre_dispatch='2*n_jobs', error_score=nan)[source]#
Evaluate a score by cross-validation.
Read more in the User Guide.
- Parameters:
- estimatorestimator object implementing ‘fit’
The object to use to fit the data.
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The data to fit. Can be for example a list, or an array.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs), default=None
The target variable to try to predict in the case of supervised learning.
- groupsarray-like of shape (n_samples,), default=None
Group labels for the samples used while splitting the dataset into train/test set. Only used in conjunction with a “Group” cv instance (e.g.,
GroupKFold).Changed in version 1.4:
groupscan only be passed if metadata routing is not enabled viasklearn.set_config(enable_metadata_routing=True). When routing is enabled, passgroupsalongside other metadata via theparamsargument instead. E.g.:cross_val_score(..., params={'groups': groups}).- scoringstr or callable, default=None
A str (see model evaluation documentation) or a scorer callable object / function with signature
scorer(estimator, X, y)which should return only a single value.Similar to
cross_validatebut only a single metric is permitted.If
None, the estimator’s default scorer (if available) is used.- cvint, cross-validation generator or an iterable, default=None
Determines the cross-validation splitting strategy. Possible inputs for cv are:
None, to use the default 5-fold cross validation,int, to specify the number of folds in a
(Stratified)KFold,An iterable that generates (train, test) splits as arrays of indices.
For
int/Noneinputs, if the estimator is a classifier andyis either binary or multiclass,StratifiedKFoldis used. In all other cases,KFoldis used. These splitters are instantiated withshuffle=Falseso the splits will be the same across calls.Refer User Guide for the various cross-validation strategies that can be used here.
Changed in version 0.22:
cvdefault value ifNonechanged from 3-fold to 5-fold.- n_jobsint, default=None
Number of jobs to run in parallel. Training the estimator and computing the score are parallelized over the cross-validation splits.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- verboseint, default=0
The verbosity level.
- fit_paramsdict, default=None
Parameters to pass to the fit method of the estimator.
Deprecated since version 1.4: This parameter is deprecated and will be removed in version 1.6. Use
paramsinstead.- paramsdict, default=None
Parameters to pass to the underlying estimator’s
fit, the scorer, and the CV splitter.New in version 1.4.
- pre_dispatchint or str, default=’2*n_jobs’
Controls the number of jobs that get dispatched during parallel execution. Reducing this number can be useful to avoid an explosion of memory consumption when more jobs get dispatched than CPUs can process. This parameter can be:
None, in which case all the jobs are immediately created and spawned. Use this for lightweight and fast-running jobs, to avoid delays due to on-demand spawning of the jobsAn int, giving the exact number of total jobs that are spawned
A str, giving an expression as a function of n_jobs, as in ‘2*n_jobs’
- error_score‘raise’ or numeric, default=np.nan
Value to assign to the score if an error occurs in estimator fitting. If set to ‘raise’, the error is raised. If a numeric value is given, FitFailedWarning is raised.
New in version 0.20.
- Returns:
- scoresndarray of float of shape=(len(list(cv)),)
Array of scores of the estimator for each run of the cross validation.
See also
cross_validateTo run cross-validation on multiple metrics and also to return train scores, fit times and score times.
cross_val_predictGet predictions from each split of cross-validation for diagnostic purposes.
sklearn.metrics.make_scorerMake a scorer from a performance metric or loss function.
Examples
>>> from sklearn import datasets, linear_model >>> from sklearn.model_selection import cross_val_score >>> diabetes = datasets.load_diabetes() >>> X = diabetes.data[:150] >>> y = diabetes.target[:150] >>> lasso = linear_model.Lasso() >>> print(cross_val_score(lasso, X, y, cv=3)) [0.3315057 0.08022103 0.03531816]
Examples using sklearn.model_selection.cross_val_score#
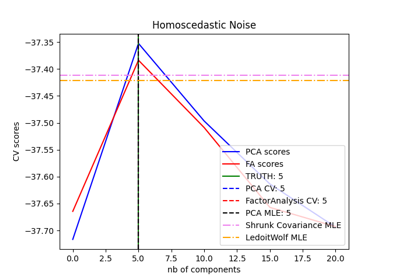
Model selection with Probabilistic PCA and Factor Analysis (FA)
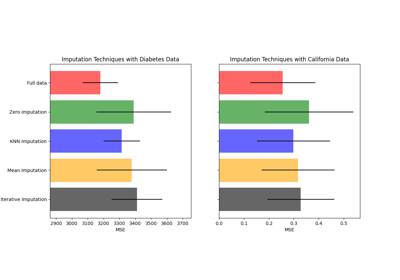
Imputing missing values before building an estimator
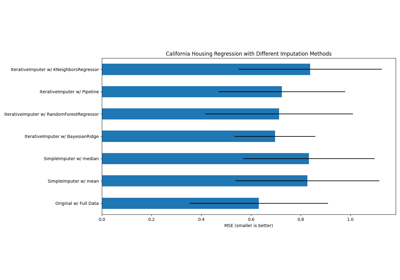
Imputing missing values with variants of IterativeImputer