sklearn.datasets.make_classification#
- sklearn.datasets.make_classification(n_samples=100, n_features=20, *, n_informative=2, n_redundant=2, n_repeated=0, n_classes=2, n_clusters_per_class=2, weights=None, flip_y=0.01, class_sep=1.0, hypercube=True, shift=0.0, scale=1.0, shuffle=True, random_state=None)[source]#
Generate a random n-class classification problem.
This initially creates clusters of points normally distributed (std=1) about vertices of an
n_informative-dimensional hypercube with sides of length2*class_sepand assigns an equal number of clusters to each class. It introduces interdependence between these features and adds various types of further noise to the data.Without shuffling,
Xhorizontally stacks features in the following order: the primaryn_informativefeatures, followed byn_redundantlinear combinations of the informative features, followed byn_repeatedduplicates, drawn randomly with replacement from the informative and redundant features. The remaining features are filled with random noise. Thus, without shuffling, all useful features are contained in the columnsX[:, :n_informative + n_redundant + n_repeated].For an example of usage, see Plot randomly generated classification dataset.
Read more in the User Guide.
- Parameters:
- n_samplesint, default=100
The number of samples.
- n_featuresint, default=20
The total number of features. These comprise
n_informativeinformative features,n_redundantredundant features,n_repeatedduplicated features andn_features-n_informative-n_redundant-n_repeateduseless features drawn at random.- n_informativeint, default=2
The number of informative features. Each class is composed of a number of gaussian clusters each located around the vertices of a hypercube in a subspace of dimension
n_informative. For each cluster, informative features are drawn independently from N(0, 1) and then randomly linearly combined within each cluster in order to add covariance. The clusters are then placed on the vertices of the hypercube.- n_redundantint, default=2
The number of redundant features. These features are generated as random linear combinations of the informative features.
- n_repeatedint, default=0
The number of duplicated features, drawn randomly from the informative and the redundant features.
- n_classesint, default=2
The number of classes (or labels) of the classification problem.
- n_clusters_per_classint, default=2
The number of clusters per class.
- weightsarray-like of shape (n_classes,) or (n_classes - 1,), default=None
The proportions of samples assigned to each class. If None, then classes are balanced. Note that if
len(weights) == n_classes - 1, then the last class weight is automatically inferred. More thann_samplessamples may be returned if the sum ofweightsexceeds 1. Note that the actual class proportions will not exactly matchweightswhenflip_yisn’t 0.- flip_yfloat, default=0.01
The fraction of samples whose class is assigned randomly. Larger values introduce noise in the labels and make the classification task harder. Note that the default setting flip_y > 0 might lead to less than
n_classesin y in some cases.- class_sepfloat, default=1.0
The factor multiplying the hypercube size. Larger values spread out the clusters/classes and make the classification task easier.
- hypercubebool, default=True
If True, the clusters are put on the vertices of a hypercube. If False, the clusters are put on the vertices of a random polytope.
- shiftfloat, ndarray of shape (n_features,) or None, default=0.0
Shift features by the specified value. If None, then features are shifted by a random value drawn in [-class_sep, class_sep].
- scalefloat, ndarray of shape (n_features,) or None, default=1.0
Multiply features by the specified value. If None, then features are scaled by a random value drawn in [1, 100]. Note that scaling happens after shifting.
- shufflebool, default=True
Shuffle the samples and the features.
- random_stateint, RandomState instance or None, default=None
Determines random number generation for dataset creation. Pass an int for reproducible output across multiple function calls. See Glossary.
- Returns:
- Xndarray of shape (n_samples, n_features)
The generated samples.
- yndarray of shape (n_samples,)
The integer labels for class membership of each sample.
See also
make_blobsSimplified variant.
make_multilabel_classificationUnrelated generator for multilabel tasks.
Notes
The algorithm is adapted from Guyon [1] and was designed to generate the “Madelon” dataset.
References
[1]I. Guyon, “Design of experiments for the NIPS 2003 variable selection benchmark”, 2003.
Examples
>>> from sklearn.datasets import make_classification >>> X, y = make_classification(random_state=42) >>> X.shape (100, 20) >>> y.shape (100,) >>> list(y[:5]) [0, 0, 1, 1, 0]
Examples using sklearn.datasets.make_classification#
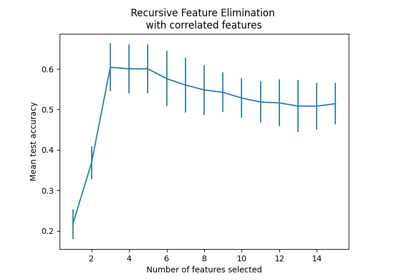
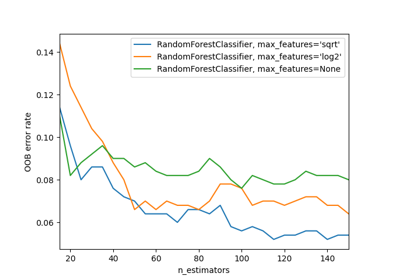
Recursive feature elimination with cross-validation
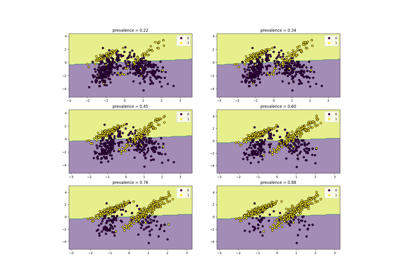
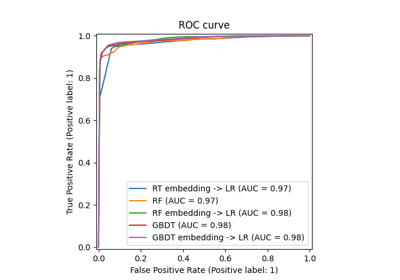
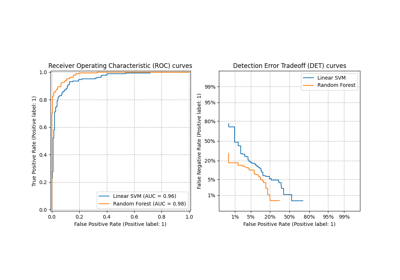
Class Likelihood Ratios to measure classification performance
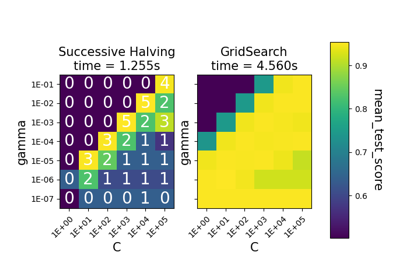
Comparison between grid search and successive halving