sklearn.impute.IterativeImputer#
- class sklearn.impute.IterativeImputer(estimator=None, *, missing_values=nan, sample_posterior=False, max_iter=10, tol=0.001, n_nearest_features=None, initial_strategy='mean', fill_value=None, imputation_order='ascending', skip_complete=False, min_value=-inf, max_value=inf, verbose=0, random_state=None, add_indicator=False, keep_empty_features=False)[source]#
Multivariate imputer that estimates each feature from all the others.
A strategy for imputing missing values by modeling each feature with missing values as a function of other features in a round-robin fashion.
Read more in the User Guide.
New in version 0.21.
Note
This estimator is still experimental for now: the predictions and the API might change without any deprecation cycle. To use it, you need to explicitly import
enable_iterative_imputer:>>> # explicitly require this experimental feature >>> from sklearn.experimental import enable_iterative_imputer # noqa >>> # now you can import normally from sklearn.impute >>> from sklearn.impute import IterativeImputer
- Parameters:
- estimatorestimator object, default=BayesianRidge()
The estimator to use at each step of the round-robin imputation. If
sample_posterior=True, the estimator must supportreturn_stdin itspredictmethod.- missing_valuesint or np.nan, default=np.nan
The placeholder for the missing values. All occurrences of
missing_valueswill be imputed. For pandas’ dataframes with nullable integer dtypes with missing values,missing_valuesshould be set tonp.nan, sincepd.NAwill be converted tonp.nan.- sample_posteriorbool, default=False
Whether to sample from the (Gaussian) predictive posterior of the fitted estimator for each imputation. Estimator must support
return_stdin itspredictmethod if set toTrue. Set toTrueif usingIterativeImputerfor multiple imputations.- max_iterint, default=10
Maximum number of imputation rounds to perform before returning the imputations computed during the final round. A round is a single imputation of each feature with missing values. The stopping criterion is met once
max(abs(X_t - X_{t-1}))/max(abs(X[known_vals])) < tol, whereX_tisXat iterationt. Note that early stopping is only applied ifsample_posterior=False.- tolfloat, default=1e-3
Tolerance of the stopping condition.
- n_nearest_featuresint, default=None
Number of other features to use to estimate the missing values of each feature column. Nearness between features is measured using the absolute correlation coefficient between each feature pair (after initial imputation). To ensure coverage of features throughout the imputation process, the neighbor features are not necessarily nearest, but are drawn with probability proportional to correlation for each imputed target feature. Can provide significant speed-up when the number of features is huge. If
None, all features will be used.- initial_strategy{‘mean’, ‘median’, ‘most_frequent’, ‘constant’}, default=’mean’
Which strategy to use to initialize the missing values. Same as the
strategyparameter inSimpleImputer.- fill_valuestr or numerical value, default=None
When
strategy="constant",fill_valueis used to replace all occurrences of missing_values. For string or object data types,fill_valuemust be a string. IfNone,fill_valuewill be 0 when imputing numerical data and “missing_value” for strings or object data types.New in version 1.3.
- imputation_order{‘ascending’, ‘descending’, ‘roman’, ‘arabic’, ‘random’}, default=’ascending’
The order in which the features will be imputed. Possible values:
'ascending': From features with fewest missing values to most.'descending': From features with most missing values to fewest.'roman': Left to right.'arabic': Right to left.'random': A random order for each round.
- skip_completebool, default=False
If
Truethen features with missing values duringtransformwhich did not have any missing values duringfitwill be imputed with the initial imputation method only. Set toTrueif you have many features with no missing values at bothfitandtransformtime to save compute.- min_valuefloat or array-like of shape (n_features,), default=-np.inf
Minimum possible imputed value. Broadcast to shape
(n_features,)if scalar. If array-like, expects shape(n_features,), one min value for each feature. The default is-np.inf.Changed in version 0.23: Added support for array-like.
- max_valuefloat or array-like of shape (n_features,), default=np.inf
Maximum possible imputed value. Broadcast to shape
(n_features,)if scalar. If array-like, expects shape(n_features,), one max value for each feature. The default isnp.inf.Changed in version 0.23: Added support for array-like.
- verboseint, default=0
Verbosity flag, controls the debug messages that are issued as functions are evaluated. The higher, the more verbose. Can be 0, 1, or 2.
- random_stateint, RandomState instance or None, default=None
The seed of the pseudo random number generator to use. Randomizes selection of estimator features if
n_nearest_featuresis notNone, theimputation_orderifrandom, and the sampling from posterior ifsample_posterior=True. Use an integer for determinism. See the Glossary.- add_indicatorbool, default=False
If
True, aMissingIndicatortransform will stack onto output of the imputer’s transform. This allows a predictive estimator to account for missingness despite imputation. If a feature has no missing values at fit/train time, the feature won’t appear on the missing indicator even if there are missing values at transform/test time.- keep_empty_featuresbool, default=False
If True, features that consist exclusively of missing values when
fitis called are returned in results whentransformis called. The imputed value is always0except wheninitial_strategy="constant"in which casefill_valuewill be used instead.New in version 1.2.
- Attributes:
- initial_imputer_object of type
SimpleImputer Imputer used to initialize the missing values.
- imputation_sequence_list of tuples
Each tuple has
(feat_idx, neighbor_feat_idx, estimator), wherefeat_idxis the current feature to be imputed,neighbor_feat_idxis the array of other features used to impute the current feature, andestimatoris the trained estimator used for the imputation. Length isself.n_features_with_missing_ * self.n_iter_.- n_iter_int
Number of iteration rounds that occurred. Will be less than
self.max_iterif early stopping criterion was reached.- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
- n_features_with_missing_int
Number of features with missing values.
- indicator_
MissingIndicator Indicator used to add binary indicators for missing values.
Noneifadd_indicator=False.- random_state_RandomState instance
RandomState instance that is generated either from a seed, the random number generator or by
np.random.
- initial_imputer_object of type
See also
SimpleImputerUnivariate imputer for completing missing values with simple strategies.
KNNImputerMultivariate imputer that estimates missing features using nearest samples.
Notes
To support imputation in inductive mode we store each feature’s estimator during the
fitphase, and predict without refitting (in order) during thetransformphase.Features which contain all missing values at
fitare discarded upontransform.Using defaults, the imputer scales in \(\mathcal{O}(knp^3\min(n,p))\) where \(k\) =
max_iter, \(n\) the number of samples and \(p\) the number of features. It thus becomes prohibitively costly when the number of features increases. Settingn_nearest_features << n_features,skip_complete=Trueor increasingtolcan help to reduce its computational cost.Depending on the nature of missing values, simple imputers can be preferable in a prediction context.
References
Examples
>>> import numpy as np >>> from sklearn.experimental import enable_iterative_imputer >>> from sklearn.impute import IterativeImputer >>> imp_mean = IterativeImputer(random_state=0) >>> imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) IterativeImputer(random_state=0) >>> X = [[np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9]] >>> imp_mean.transform(X) array([[ 6.9584..., 2. , 3. ], [ 4. , 2.6000..., 6. ], [10. , 4.9999..., 9. ]])
For a more detailed example see Imputing missing values before building an estimator or Imputing missing values with variants of IterativeImputer.
Methods
fit(X[, y])Fit the imputer on
Xand return self.fit_transform(X[, y])Fit the imputer on
Xand return the transformedX.get_feature_names_out([input_features])Get output feature names for transformation.
Raise
NotImplementedError.get_params([deep])Get parameters for this estimator.
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Impute all missing values in
X.- fit(X, y=None)[source]#
Fit the imputer on
Xand return self.- Parameters:
- Xarray-like, shape (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.- yIgnored
Not used, present for API consistency by convention.
- Returns:
- selfobject
Fitted estimator.
- fit_transform(X, y=None)[source]#
Fit the imputer on
Xand return the transformedX.- Parameters:
- Xarray-like, shape (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.- yIgnored
Not used, present for API consistency by convention.
- Returns:
- Xtarray-like, shape (n_samples, n_features)
The imputed input data.
- get_feature_names_out(input_features=None)[source]#
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names.
- get_metadata_routing()[source]#
Raise
NotImplementedError.This estimator does not support metadata routing yet.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- set_output(*, transform=None)[source]#
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame output"polars": Polars outputNone: Transform configuration is unchanged
New in version 1.4:
"polars"option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- transform(X)[source]#
Impute all missing values in
X.Note that this is stochastic, and that if
random_stateis not fixed, repeated calls, or permuted input, results will differ.- Parameters:
- Xarray-like of shape (n_samples, n_features)
The input data to complete.
- Returns:
- Xtarray-like, shape (n_samples, n_features)
The imputed input data.
Examples using sklearn.impute.IterativeImputer#
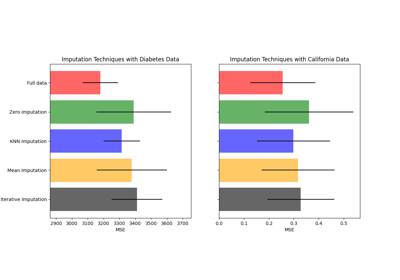
Imputing missing values before building an estimator
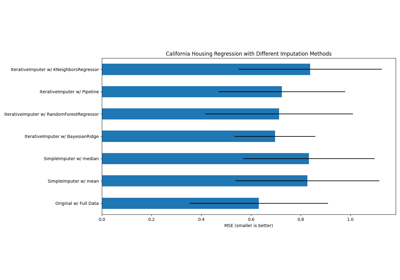
Imputing missing values with variants of IterativeImputer
